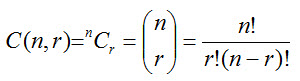Methods
Combinations
Copyright (C) 2011 Jennifer D. Warrender, Newcastle Univeristy
MSA Construction
For the creating of the alignments, combinations that do not allow repetition were used. The mathematical formula for the number of combinations is as follows (Pólya, et al., 2010):
Using the formula, the number of MSAs for each subset size could be calculated.
In order to determine generic rules used to obtain the best alignments for the PON tools; it was required to create every possible MSA size and combination of the sequences input. These subsets were created using a modified CombinationGenerator program based on an online java class that created combinations of the sequences, excluding H. sapiens as the H. sapiens sequence needs to be found at the top of every alignment for the PON tools - as human mutations are the only concern. Thus each alignment had human on top as the query and n other sequences where n is greater than 1. This resulted in different MSAs to be run in conjunction with the PON tools.
Optimization of parameters for the assessment of Unclassified Disease Gene Sequence Variants